I recently wrote a post about the ENCODE program that was going on. I had posted an article where it showed that geneticists have recently made a startling discovery showing that the genome and the few genes in it was only a small piece in a far bigger design that was just revealed to the researchers. I was very much mistaken (and very wrong) on the amount of knowledge of the real professional geneticists who do work in the subject. The press, the journalists, and the writers are mostly in the dark on the subject of genetics like most of the general public including me.
Let me clarify that my understanding of genetics is still at a very rudimentary level that is from 1 class in college with biochemistry and another class on basic biology. I have never taken a class on genetics, although I have sat in on one for 2 whole classes.
I always want to mention to the reader that this website is not a place for me to spew out the vast encyclopedia of height increase knowledge and information I have learned in the last 5 years. This website is a project, an on going , always evolving piece of electronic knowledge database that will be corrected when a mistake is made. I have already had to issue public apologies to organizations because I made a wrong judgement of intent.
I am learning as time goes on and I am continuously adding more and more information to my brain and this site. If you have read the most recent articles you would have noted that most of them were posts to help also myself understand the mechanics of human growth better. You can actually watch the changes being made.
This article copied and posted below was taken from the website Ars Technica HERE and written by John Timmer
Most of what you read was wrong: how press releases rewrote scientific history
Repeating myths may make good stories, but it breeds confusion. See the ENCODE news.
This was more than a matter of semantics. Many press reports that resulted painted an entirely fictitious history of biology’s past, along with a misleading picture of its present. As a result, the public that relied on those press reports now has a completely mistaken view of our current state of knowledge (this happens to be the exact opposite of what journalism is intended to accomplish). But you can’t entirely blame the press in this case. They were egged on by the journals and university press offices that promoted the work—and, in some cases, the scientists themselves.
To understand why, we’ll need a bit of biology and a bit of history before we can turn back to the latest results and the public response to them.
What we know about DNA, and when we knew it
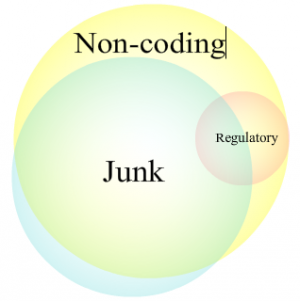
Among other things, DNA has at least two key functions. First, it codes for the proteins that perform most of a cell’s functions. Second, it has control sequences that don’t encode anything, but determine when and where the coding sequences are active. We’ve had some indication that non-coding DNA played key regulatory roles since the 1960s, when the Lac operon was described and won its discoverers the Nobel Prize.
The Lac operon is present in bacterial genomes, which are under extreme pressure to carry as little DNA as possible. The typical bacterial genome is over 85 percent protein-coding DNA, leaving just a small fraction for regulatory purposes. But that isn’t generally true of vertebrates.
The coding portions of vertebrate genes turned out to be interrupted by noncoding regions, called introns. Some of these are huge—roughly a third the size of some of the smaller bacterial genomes. Vertebrate genomes also appeared to be littered with old and disabled viruses and mobile genetic parasites called transposons. Even some of the coding portions seemed a bit useless—near exact duplicates of genes were common, as were mutated and disabled copies. Many of these apparently useless pieces of DNA continued to carry sites for regulatory DNA binding proteins and continued to make RNA.
(To give you an idea of how mainstream all this was, I spent some time working on a mouse gene that was thought to be superfluous because it was a near-exact copy of a gene used by the immune system. But the copy was only expressed in males because a mobile genetic element’s regulatory sequences had been inserted nearby. And I knew all this as an undergrad in the late 1980s).
By the time we sequenced the human genome, we discovered that this seemingly useless stuff was the majority. Over half the genome was built from the remains of viruses and transposons. Introns accounted for another large fraction. And all of it seemed to be an evolutionary accident. One fish, the fugu, lacks a lot of this DNA, and seems to get along fine, while many salamanders have ten times the DNA per cell that humans do. And if you looked at the DNA of different mammals, the vast majority of it (about 95 percent) wasn’t shared by different species.
These findings seemed to support a model that was first proposed back in the 1970s, which picked up the (possibly unfortunate) moniker junk DNA. Genomic accidents—duplicating genes, picking up a virus—happen at a steady rate. Individually, these don’t cause an appreciable cost in terms of fitness, so species aren’t under a strong selective pressure to get rid of it, and pieces could linger in the genome for millions of years. But the typical bit of junk doesn’t do anything positive for the animals that carry it.
You could even consider the idea of junk DNA to be a scientific hypothesis. It notes that animal genomes experience several processes that produce superfluous bits of DNA, predicts that these will not cause enough harm to be selected for elimination, and proposes an outcome: genomes littered with random bits of history that have no impact on an organism’s fitness.
Junk dies a thousand deaths
For decades we’ve known a few things: some pieces of non-coding DNA were critically important, since they controlled when and where the coding pieces were used; but there was a lot of other non-coding DNA and a good hypothesis, junk DNA, to explain why it was there.
Unfortunately, things like well-established facts make for a lousy story. So instead, the press has often turned to myths, aided and abetted by the university press offices and scientists that should have been helping to make sure they produced an accurate story.
Discovery of new regulatory DNA isn’t usually surprising, given that we’ve known it’s out there for decades. There has been a steady stream of press releases that act as if finding a function for non-coding DNA is a complete surprise. And many of these are accompanied by quotes from scientists that support this false narrative.
The same thing goes for junk DNA. We’ve known for decades that some individual pieces of junk DNA do something useful. Introns can regulate gene expression. Bits of former virus or transposon have been found incorporated into genes or used to regulate their expression. So some junk DNA can be useful, in much the same way that a junk yard can be a valuable source of spare parts.
But it’s important to keep these in perspective. Even if a function is assigned to a piece of junk that’s 1,000 base pairs long, that only accounts for about 1/2,250,000 of the total junk that is estimated to reside in the human genome. Put another way, it’s important not to fall into the logical fallacy that finding a use for one piece of junk must mean that all of it is useful.
Despite that, many new findings in this area are accompanied by some variation on the declaration that junk is dead. Both press officers and scientists have presented a single useful piece of virus as definitively establishing that every virus, transposon, and dead gene in the human genome is essential for our collective health and survival.
A bad precedent, repeated
This brings us to the ENCODE project, which was set up to provide a comprehensive look at how the human genome behaves inside cells. Back in 2007, the consortium published its first results after having surveyed one percent of the human genome, and the results foreshadowed this past week’s events. The first work largely looked at what parts of the genome were made into RNA, a key carrier of genetic information. But the ENCODE press materials performed a sleight-of-hand, indicating that anything made into RNA must have a noticeable impact on the organism: “the genome contains very little unused sequences; genes are just one of many types of DNA sequences that have a functional impact.”
There was a small problem with this: we already knew it probably wasn’t true. Transposons and dead viruses both produce RNAs that have no known function, and may be harmful in some contexts. So do copies of genes that are mutated into uselessness. If that weren’t enough, just a few weeks later, researchers reported that genes that are otherwise shut down often produce short pieces of RNA that are then immediately digested.
So even as the paper was released, we already knew the ENCODE definition of “functional impact” was, at best, broad to the point of being meaningless. At worst, it was actively misleading.
But because these releases are such an important part of framing the discussion that follows in the popular press, the resulting coverage reflected ENCODE’s spin on its results. If it was functional, it couldn’t be junk. The concept of junk DNA was declared dead far and wide, all based on a set of findings that were perfectly consistent with it.
Four years later, ENCODE apparently decided to kill it again.
A new definition, the same problem
In the lead paper of a series of 30 released this week, the ENCODE team decided to redefine “functional.” Instead of RNA, its new definition was more DNA focused, and included sequences that display “a reproducible biochemical signature (for example, protein binding, or a specific chromatin structure).” In other words, if a protein sticks there or the DNA isn’t packaged too tightly to be used, then it was functional.
That definition nicely encompasses the valuable regulatory DNA, which controls nearby genes through the proteins that stick to it. But—and this is critical—it also encompasses junk DNA. Viruses and transposons have regulatory DNA to ensure they’re active; genes can pick up mutations in their coding sequence that leave their regulatory DNA intact. In short, junk DNA would be expected to include some regulatory DNA, and thus appear functional by ENCODE’s definition.
By using that definition, the ENCODE project has essentially termed junk DNA functional by fiat—for the second time. And perhaps more significantly, they did so two paragraphs after declaring that their results showed 80 percent of the genome was functional.
The response has been about as unfortunate as you might predict. The Nature press materials announcing the paper proclaim, “Far from being junk, the vast majority of our DNA participates in at least one biochemical event in at least one cell type.” The European Molecular Biology Laboratory’s release said that the work “revealed that much of what has been called ‘junk DNA’ in the human genome is actually a massive control panel with millions of switches regulating the activity of our genes.” MIT’s was entitled “Researchers identify biochemical functions for most of the human genome.”
But this can’t simply be blamed on the PR staff. Scientists from the National Human Genome Research Institute have been fostering the confusion. One of its program directors was quoted by UC Santa Cruz as suggesting we thought regulatory DNA was junk: “Far from being ‘junk’ DNA, this regulatory DNA clearly makes important contributions to human disease.” And its director, Eric Green, was quoted by Penn State as saying, “we can now say that there is very little, if any, junk DNA.”
This confusion leaked over to the popular press. Bloomberg’s coverage, for example, suggests we’ve never discovered regulatory DNA: “Scientists previously thought that only genes, small pieces of DNA that make up about 1 percent of the genome, have a function.” The New York Times defined junk as “parts of the DNA that are not actual genes containing instructions for proteins.” Beyond that sort of fundamental confusion, almost every report in the mainstream press mentioned the 80 percent functional figure, but none I saw spent time providing the key context about how functional had been defined.
The part in which we conclude
ENCODE remains a phenomenally successful effort, one that will continue to pay dividends by accelerating basic science research for decades to come. And the issue of what constitutes junk DNA is likely to remain controversial—I expect we’ll continue to find more individual pieces of it that perform useful functions, but the majority will remain evolutionary baggage that doesn’t do enough harm for us to eliminate it. Since neither issue is likely to go away, it would probably be worth our time to consider how we might prevent a mess like this from happening again.
The ENCODE team itself bears a particular responsibility here. The scientists themselves should have known what the most critical part of the story was—the definition of “functional” and all the nuance and caveats involved in that—and made sure the press officers understood it. Those press officers knew they would play a key role in shaping the resulting coverage, and should have made sure they got this right. The team has now failed to do this twice.
More generally, the differences among non-coding DNA, regulatory DNA, and junk DNA aren’t really that hard to get straight. And there’s no excuse for pretending that things we’ve known for decades are a complete surprise.
Unfortunately, this is a case where scientists themselves get these details wrong very often, and their mistakes have been magnified by the press releases and coverage that has resulted. That makes it much more likely that any future coverage of these topics will repeat the past errors.
If the confused coverage of ENCODE has done anything positive, it has provoked a public response by a number of scientists. Their criticisms may help convince their colleagues to be more circumspect in the future. And maybe a few more reporters will be aware that this is an area of genuine controversy, and it will help them identify a few of the scientists they should be talking to when covering it in the future.
It’s just a shame the public had to be badly misled in the process.
Pingback: Complete List Of Posts - |